Overview
The molecular processing pipeline in HoneyBee handles various types of molecular data, including DNA methylation, gene expression, protein expression, DNA mutation, and miRNA expression. The pipeline preprocesses, integrates, and generates embeddings from these diverse molecular profiles for downstream machine learning applications.
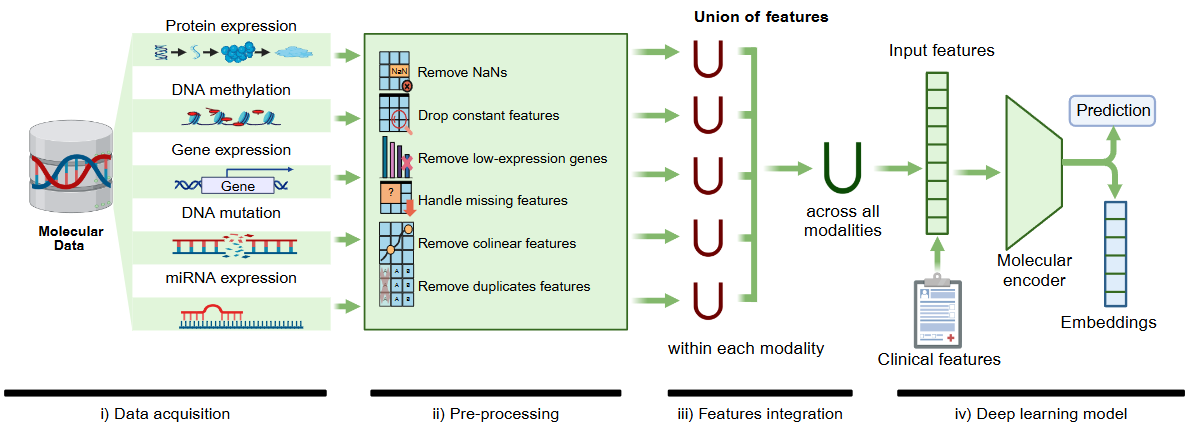
Note: The molecular processing capabilities described below are currently implemented through the SeNMo framework and operate as an independent component. We are actively working to integrate these functionalities directly into the HoneyBee ecosystem in upcoming releases to provide a more unified and streamlined experience.
